Improved data-analytical tools are needed to. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis.

Orchestrating Microbiome Analysis
In addition to creating indels or knockouts scientists can encourage a precise form of repair homology-directed repair.

. Meta-analysis of gut microbiome studies identifies disease-specific and. The tutorial walkthrough of the DADA2 pipeline on paired end Illumina Miseq data. The data are extremely noisy and large p and small n.
2011Lets find this experiment in the platform and open it in Metainfo Editor. Grosman Kaplan K Anglin R et al. Due to its division of X into Xˆ and Xˆ o OPLS splits R 2 X into R 2 X p and R 2 X o the explained sum of squares of the Y-predictive and Y-uncorrelated components of X respectively.
Yinglin Xia in Progress in Molecular Biology and Translational Science 2020. These studies provide the essential material to deeply explore host-microbiome associations and their relation to the development and progression of various complex diseases. Bioconductor workflow for microbiome data analysis.
In the field of microbial community ecology many packages can be used for the statistical analysis such as vegan Oksanen et al. The commonly amplified ITS1 and ITS2 regions range from 200 - 600 bp in length. The gut microbiome and inflammation in obsessive-compulsive disorder.
For todays tutorial we will use public data from the QIIME2 website mkdir qiime2-moving-pictures-tutorial cd qiime2-moving-pictures-tutorial. 2010However with the development of. How to perform a meta-analysis with R.
Setting up an exome sequencing experiment. First off lets choose exome sequencing data. A Hands-On Tutorial Amanda Birmingham Center for Computational Biology Bioinformatics University of California at San Diego.
The dada2 R package manual. You can upload your own data using Import button or search through all public experiments we have on the platform. The authors compared the.
To assess overall correspondence between host gene regulation and gut microbiome composition in CRC IBD and IBS we performed Procrustes analysis in R using the vegan package version 24-5. Evid Based Ment Health. 7132 Independent principal component analysis IPCA.
IPCA 311 was proposed to solve the problems of both the high dimensionality of high-throughput data and noisy characteristics of biological data in omics studies. HDR by providing a DNA sequence that the cell can use as a repair template to insert knock in a matching DNA sequence into the breakExample applications include modification of a promoter sequence or gene insertion of an exogenous reporter eg. From raw reads to community analyses.
The key addition to this workflow compared to the tutorial is the identification and removal of primers from the reads and the verification of primer orientation and removal. The number of microbiome-related studies has notably increased the availability of data on human microbiome composition and function. Microbiome Analysis with QIIME2.
Here the authors present a pig integrated gene catalog and metagenome-assembled genomes which they construct from swine gut microbiomes spanning various ages sexes breeds geographical. Rücker G Schwarzer G. Highly disparate R 2 and Q 2 values ie.
Our group at the Harvard Chan Center for the Microbiome in Public Health focuses on understanding the function of microbial communities particularly that of the human microbiomeThis means learning to use the microbiome to predict disease onset progression and outcomes as well as developing ways to modify it for health maintenance and therapy. The custom functions that read external data files and return an instance of the phyloseq-class are called importersValidity and coherency between data components are checked by the phyloseq-class constructor phyloseq which is invoked internally by the importers and is also the recommended function for creating a phyloseq object from manually imported data. Chapter 1 Background R language R Core Team 2016 and its packages ecosystem are wonderful tools for data analysis.
Soil microbial diversity patterns have been well documented in a wide variety of habitats 123From an initial focus on the description of patterns attention is now turning towards the underlying processes influencing the structure of microbial communities 4 5Cropland soils represent a relatively neglected area in terms of both patterns and processes. Unlike the 16S rRNA gene the ITS region is highly variable in length. 2019 ape Paradis and Schliep 2018 and picante Kembel et al.
Omics data have the problems. 文章目录简介工作原理优势功能模块软件安装数据库配置CheckM数据库KRAKEN数据库NCBI_ntNCBI物种信息人类基因组bmt索引设置数据库位置参数简介Reference猜你喜欢写在后面 简介 MetaWRAP这是一套强大的宏基因组分析流程专注于宏基因组Binning文章于2018年9月15日发表于Microbiome. R 2 Q 2 are an indicator of possible model over-fitting in supervised analyses.
Our analysis will be based on data coming from Clark et al.
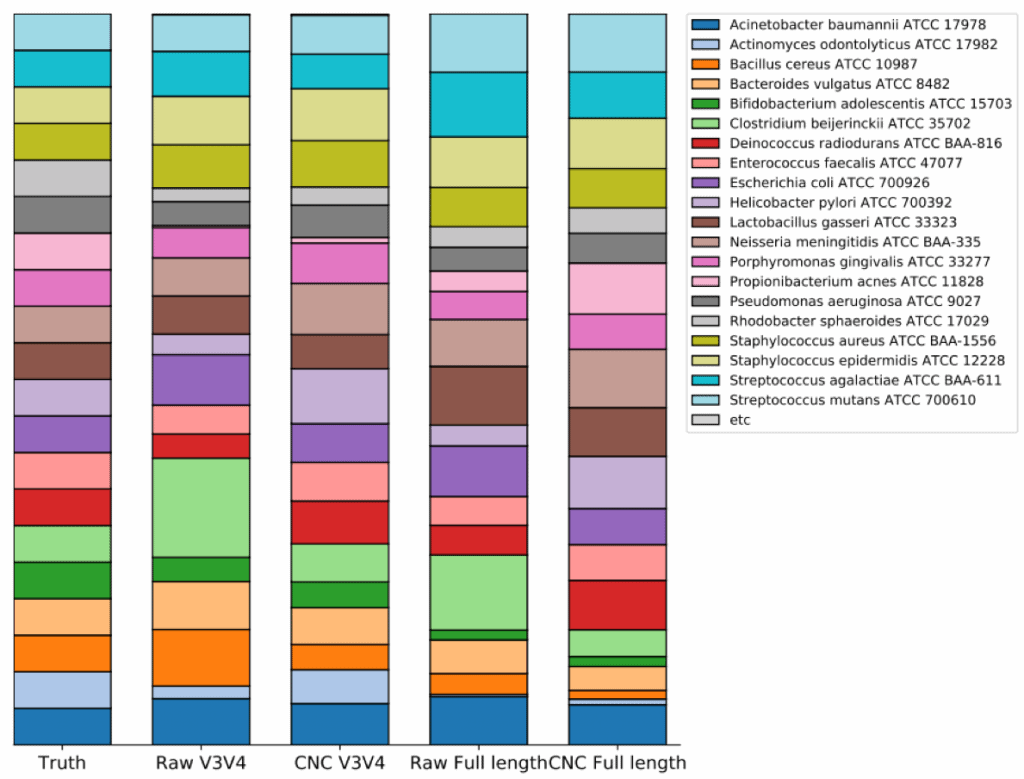
Tutorial 16s Microbiome Analysis Of Atcc R Microbiome Standards Ezbiocloud Help Center

Making Heatmaps With R For Microbiome Analysis The Molecular Ecologist
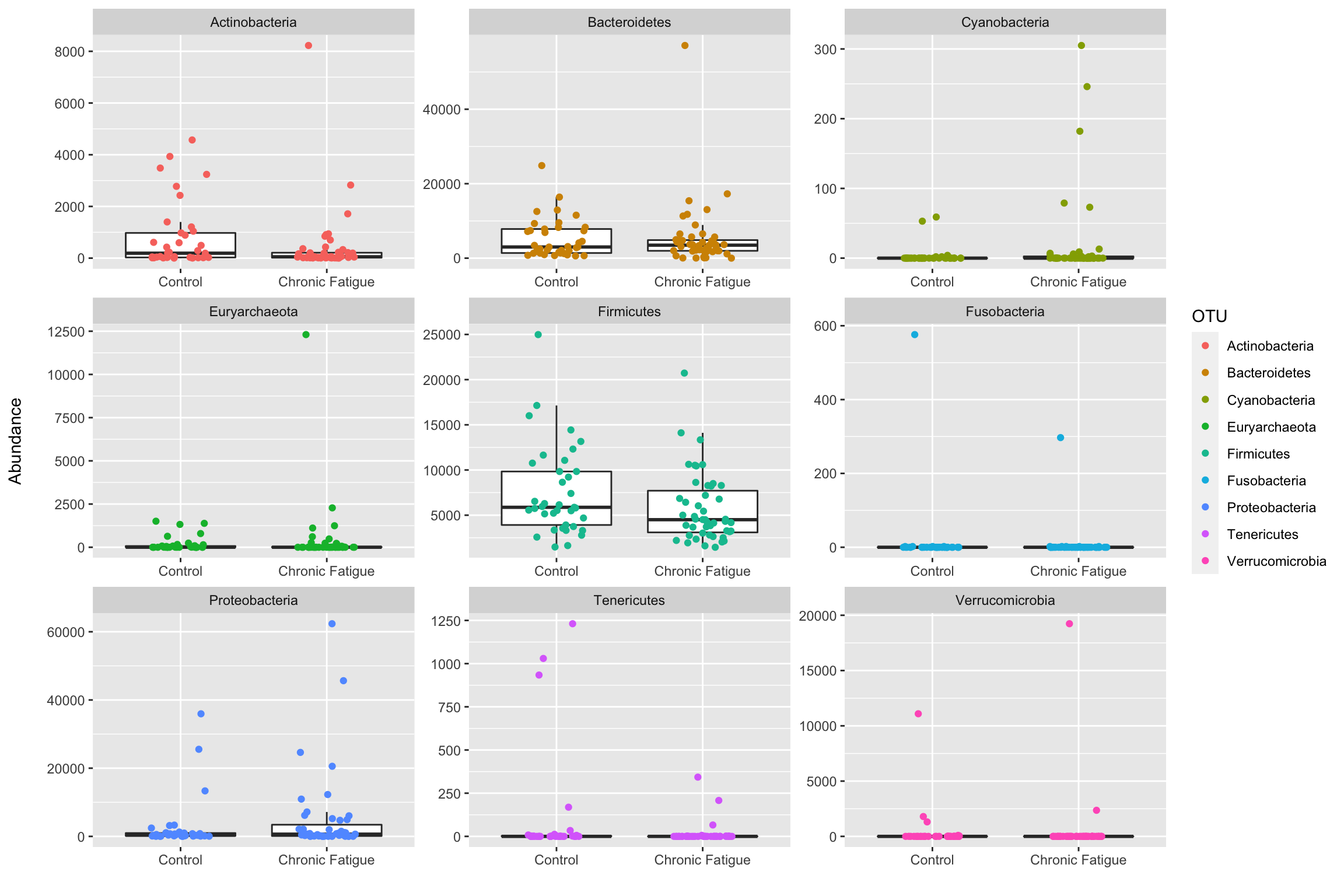
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
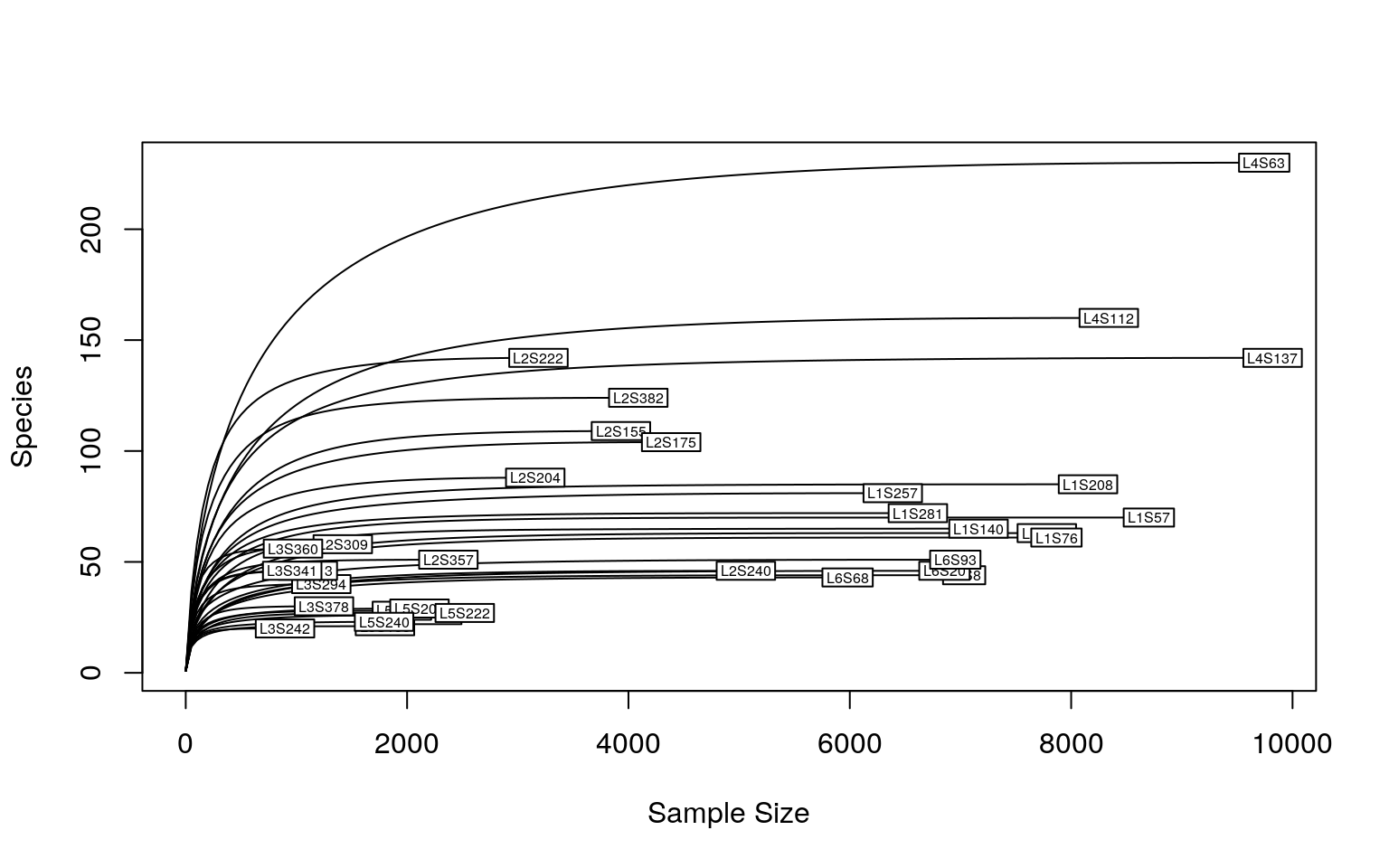
Tutorial For Microbiome Analysis In R Yan Hui
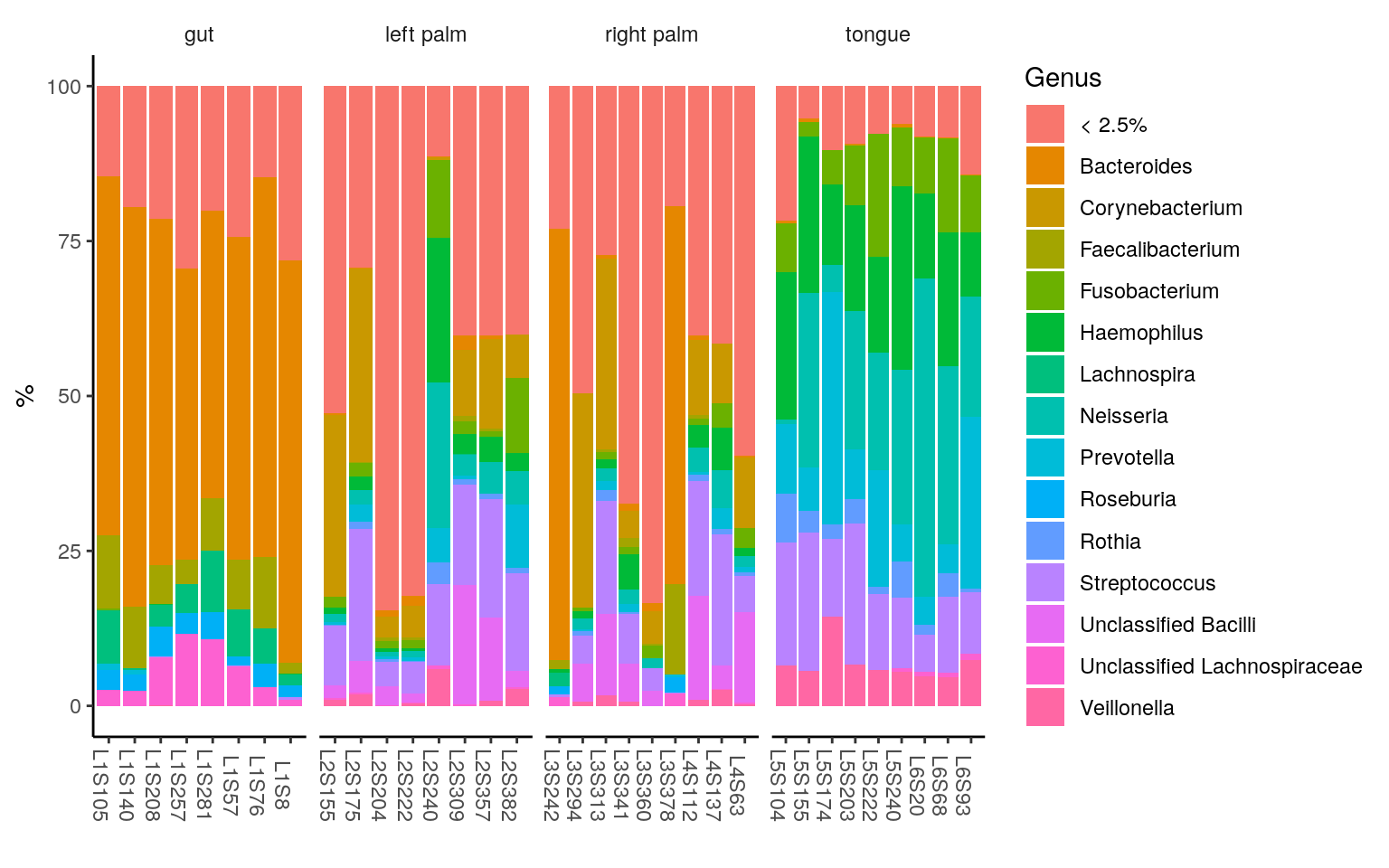
Tutorial For Microbiome Analysis In R Yan Hui
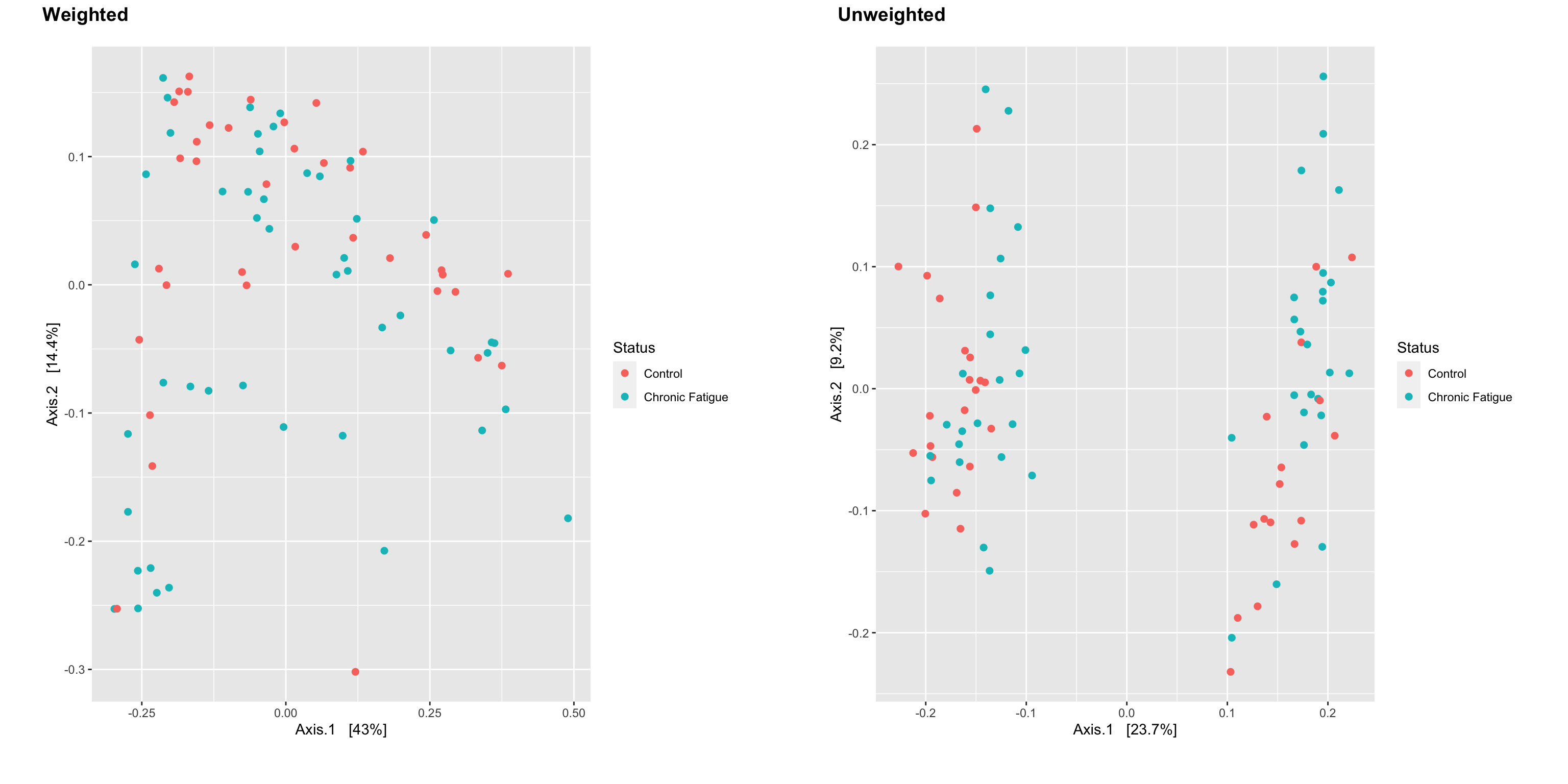
Introduction To The Statistical Analysis Of Microbiome Data In R Academic

0 comments
Post a Comment